Research in the Lab of Dr. Amy T. Hark
Welcome to the Hark Lab!
We investigate the effects of DNA packaging and genome organization on gene function.
Please visit us in Room 223, New Science Building.

Dr. Hark and students at the 2023 Lehigh Valley Molecular and Cell Biology Symposium
My research interests focus on the regulation of gene transcription in eukaryotic organisms. In particular, I am interested in how factors such as covalent modifications of histones, chromatin structure/architecture, and gene organization may act and interact to influence gene expression.
Impact of histone acetylation on plant development
Background on HATs and Histone Acetylation
One area of current research investigates the biological role of the histone acetyltransferase (HAT) enzyme GCN5 in developmental pathways in the model plant Arabidopsis thaliana. GCN5 can covalently modify histones (chromatin proteins) by catalyzing the addition of acetyl group to specific lysine residues. This modification is hypothesized to affect histone-DNA contacts or provide binding sites for other factors involved in regulating transcription (the histone code hypothesis). Misregulation of histone acetylation states has been implicated in cancer and developmental disorders.
Background on Arabidopsis
Why study chromatin modifying factors in Arabidopsis?
First, a little bit of background. Arabidopsis is a flowering plant, in the Brassica family, related to plants you are probably more familiar with such as broccoli and cauliflower. Arabidopsis has emerged as an experimental model for a number of reasons. For me, Arabidopsis also provides a model system to explore gene regulation in the context of its effects on fundamental developmental events, which is one of my areas of interest. Also, there has been a long history of the study of epigenetic effects on gene expression in plants, but relatively little is known about the roles chromatin-modifying factors such as GCN5 play.
Previous Work
In collaboration with Dr. Elizabeth McCain, we have undertaken a detailed characterization of the gcn5 mutant phenotype, as one way to help us identify specific GCN5 developmental targets. Our latest work in this area was published in a series of recent micropublications. Check out the links below for more details.
Kotak, J.*, A. Kendig*, K. Cann*, J. Shaffer*, A.T. Hark, and E.R. McCain. 2019. Disruption of the Histone Acetyltransferase GCN5 and the Transcriptional Coactivator ADA2b Affect Trichome Density in Arabidopsis thaliana. microPublication Biology. https://doi.org/10.17912/micropub.biology.000174
Trachtman, N.*, P. Sockler*, H. Caiola*, E.R. McCain, and A.T. Hark. 2019. Expression of the DELLA Repressor GAI and its Regulators SPY and SEC are Impacted by Disruption of Chromatin Modifiers. microPublication Biology. https://doi.org/10.17912/micropub.biology.000175
Hark, A.T. and E.R. McCain. 2019. The Histone Acetyltransferase GCN5 and the Transcriptional Coactivator ADA2b Affect Trichome Initiation in Arabidopsis thaliana. microPublication Biology. https://doi.org/10.17912/micropub.biology.000176
Current Work
Beginning in 2022, we plan to conduct some pilot experiments looking at long, non-coding RNAs (lncRNAs) in Arabidopsis and how they may interact with chromatin-based regulation of gene expression. One lncRNA of interest is SVALKA, whose coding sequence overlaps with the gene for CBF1, a key transcriptional regulator governing cold acclimation in plants. GCN5 also affects CBF1 expression. Might there be any crosstalk between SVALKA and GCN5?
Genomics Education Partnership
This project is part of my collaboration with the Genomics Education Partnership (GEP; thegep.org). In short, this work in comparative genomics involves using in silico (computer-based) analysis to annotate genes in Drosophila species (defining their start/stop sites, exon/intron boundaries, etc.). We have previously focused on how chromatin structure and gene structure and organization act and interact to impact gene function, which connects to my basic research interests and work of other members of the Hark Lab. More recently, we have begun applying our annotation skills to ask questions of how gene networks evolve. This collaborative research allows students to be part of a large-scale project and have their work published while increasing their knowledge of fundamental molecular biology and research tools.
Current and Recent Lab Members
There are opportunities for students to carry out research to address these and related questions in the field of molecular biology. I encourage you to contact me if you are interested in doing research in the lab, so that we may talk more about your area(s) of interest and potential projects.
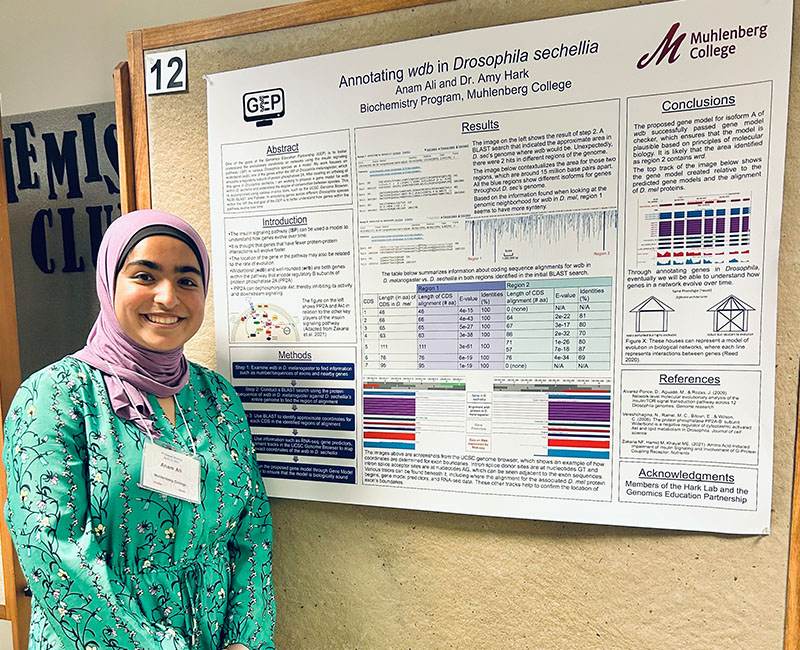
Anam Ali ’25 began working on the GEP Pathways Projects in Spring 2023.
|
Lauren Washco '24 joined the lab in Fall 2021. She has engaged in the GEP Pathways Project and is working on an educational case study that is an extension of that work. She is also planning to begin studying epigenetic effects of lncRNAs in Arabidopsis. Aviv Campbell '23 and Arielle Touitou '24 began gene annotation projects in Summer 2022.
|
|
|
Jonah Silverman '22, Sarah Raab '22, and Lindsey Harris '22 presented their research done in collaboration with the GEP at the 2022 annual meeting of the American Society for Biochemistry and Molecular Biology. |
|
|
|
|
|
Zachary Lill ’21 (below left) spearheaded our work on the GEP Pathways Project, which investigates how the insulin signaling network evolves in Drosophila species. Viet Le ’22 (below right), Benjamin Chen ’22, and Sarah Teitelbaum ’22 have also engaged in this research project. |
|
|
|
|
|
Natalie Trachtman '20 (left) and Iffat Imran '20 worked on GEP projects in Spring and Fall 2018 and continued this work in Spring 2019. Natalie engaged in work to understand how GCN5 impacts trichome initiation in plants during Summer 2019. |
|
|
|
|
|
Patrick Sockler '19 (right) and Josh S |
|
|
Sam Tener '18 sperheaded a new colaboration with Dr. Scott Woody and Dr. Rick Amasino at the University of Wisconsin, exploring possible connections between GCN5 and gibberellin signalling in plant development. Emily Chiacchiaro '19 has also joined this latter effort. Both Sam and Emily initially worked on the GEP project. |
|
|
Nate Wiggley '20 and Nina Hernandez joined the GEP group in Fall 2018. John Hanna '19 worked on both a pedagogical project and gene annotation in Fall 2018. |
|
|
Hanna Caiola '18 (bottom left) and Hannah Molk '18 (bottom right) started in the lab working on the Drosophila in silico genomics project and then transitioned to wet-lab work investigating putative GCN5 targets that impact trichome development. |
|
 |
 |
Hark Lab Alumni
Return to Dr. Hark's homepage
Page last updated December 2023





 haffer '19 joined the lab in Fall 2017 to work on the comparative genomics project and both also transitioned to the plant development project.
haffer '19 joined the lab in Fall 2017 to work on the comparative genomics project and both also transitioned to the plant development project.